INTRODUCTION
The role of prostaglandin-endoperoxide synthase 2 (PTGS2) in physiology and pathophysiology has been addressed in different studies since this enzyme is induced under several circumstances, including inflammation, tumor progression and cell survival[1-6]. Two PTGSs are present in mammalian cells: PTGS1, which is constitutively expressed in almost all tissues at low levels, playing a homeostatic role; and PTGS2, which is considered as an immediate early gene that is expressed in response to a wide array of cell challenges and stressors[7-10]. Both enzymes catalyze the same reaction, which constitutes the rate-limiting step in the biosynthesis of different prostanoids, such as several prostaglandins, via tissue specific prostaglandin synthases, thromboxane A2 and other eicosanoids[11,12]. Provision of arachidonic acid as substrate is dependent on the activation of phospholipase A2, which in turn, responds to different cell stressors connecting phospholipid hydrolysis to prostanoid synthesis[10,11,13,14] (Figure 1). Both PTGS isoforms are conserved among mammals and weight 70-75 kDa. They share more than 60% sequence homology in mammalian species and retain more than 85% identity when comparing orthologues from different species, displaying conserved regulatory and catalytic domains as depicted in Figure 2. Structural studies show that the isoleucine located at position 523 in PTGS1 is substituted by valine in PTGS2 (position 509) and this difference in hydrophobicity and size constitutes the basis for the design of selective, isoenzyme-specific hydrophobic inhibitors, such as the coxibs[15-17]. Regarding the conserved protein motifs, they include an epidermal growth factor-like domain followed by a membrane-binding region that allows positioning of the different PTGS in cytoplasmic micro-ambiances. The catalytic site of the enzyme involves two independent activities: the deoxygenation of arachidonic acid and an additional site responsible for the subsequent reduction via the peroxidase activity[18]. These domains are relevant for the subcellular localization of PTGS allowing the protein to interact with the luminal space of the endoplasmic reticulum and with the nuclear membrane. This is important to understand the activity of the enzyme since phospholipases and their targets, the phospholipids required to release arachidonic acid, are located in biological membranes[19-23]. Additionally, other free fatty acids, such as eicosanepentaenoic acid[24], docosahexaenoic acid[23], α-and γ-linolenic acid or linoleic acid can be metabolized by PTGS2 leading to molecules involved in the control of inflammation[25,26] (Figure 3). Several works described selective distribution of both PTGS isoforms in the cell, with a preferred positioning of PTGS2 near the nuclear structure. This is also pertinent for the fate of the products of the enzymes[10]. These prostanoids can be released to the extracellular milieu and exert their autocrine or paracrine actions either by the specific G protein-coupled receptor (GPCR)-coupled prostaglandin E2 (PGE2) receptor (EP) receptors[27], by diffusion or through the interaction with several transporters (i.e., the prostaglandin transporter system, the ABC cassettes, or the scavenger lipid receptor CD36[28-30]). In addition to GPCR-mediated early signaling, prostanoids may alter gene transcription after interaction with several nuclear receptors, such as the peroxisomal proliferator activated receptors (PPARs)[31-33].
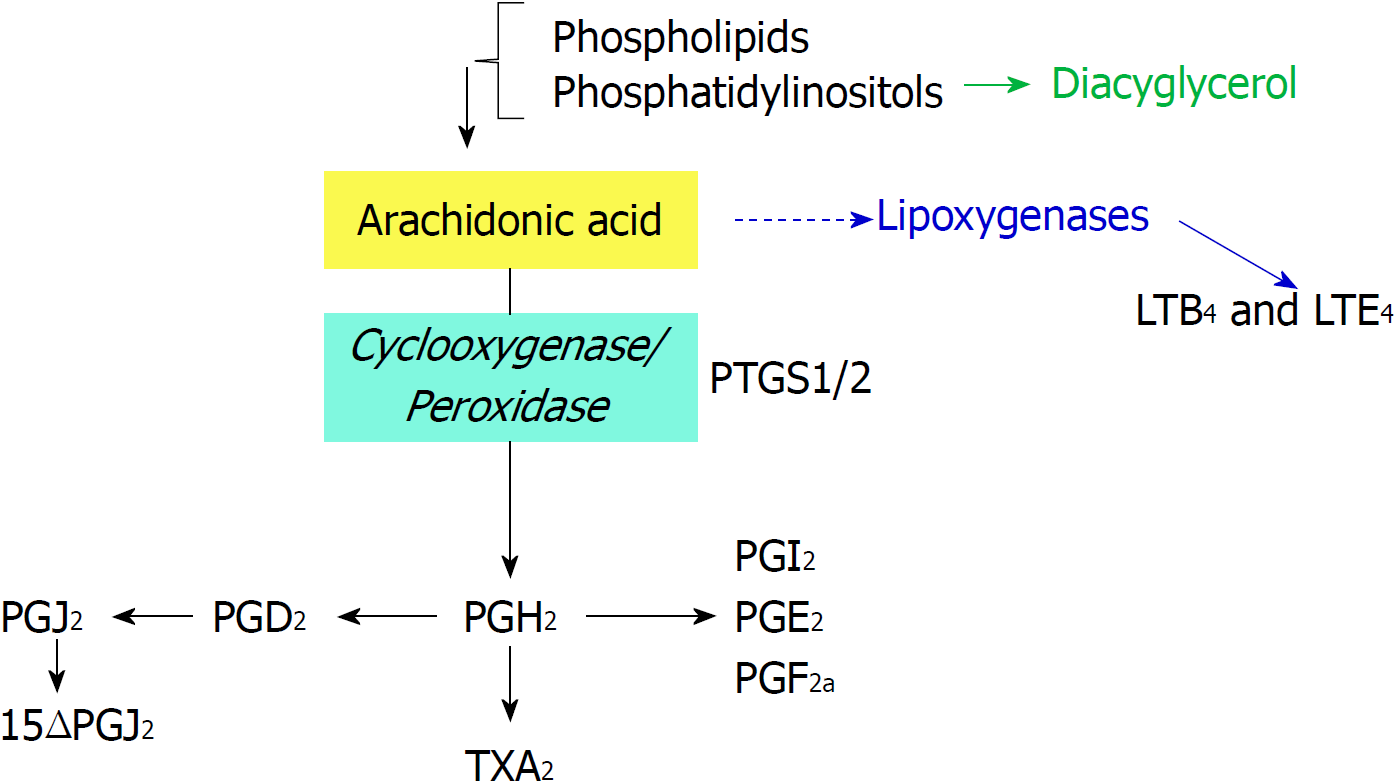
Figure 1 Biochemical reactions catalyzed by prostaglandin-endoperoxide synthase using arachidonic acid as substrate.
Phospholipids are cleaved by phospholipases to render arachidonic acid. Prostaglandin-endoperoxide synthase activity generates the precursor prostaglandin H2 that is converted in the different prostaglandins (PGs) by prostaglandin synthases and thromboxane A2, by thromboxane synthase. PG: Prostaglandin; PTGS: Prostaglandin-endoperoxide synthase.
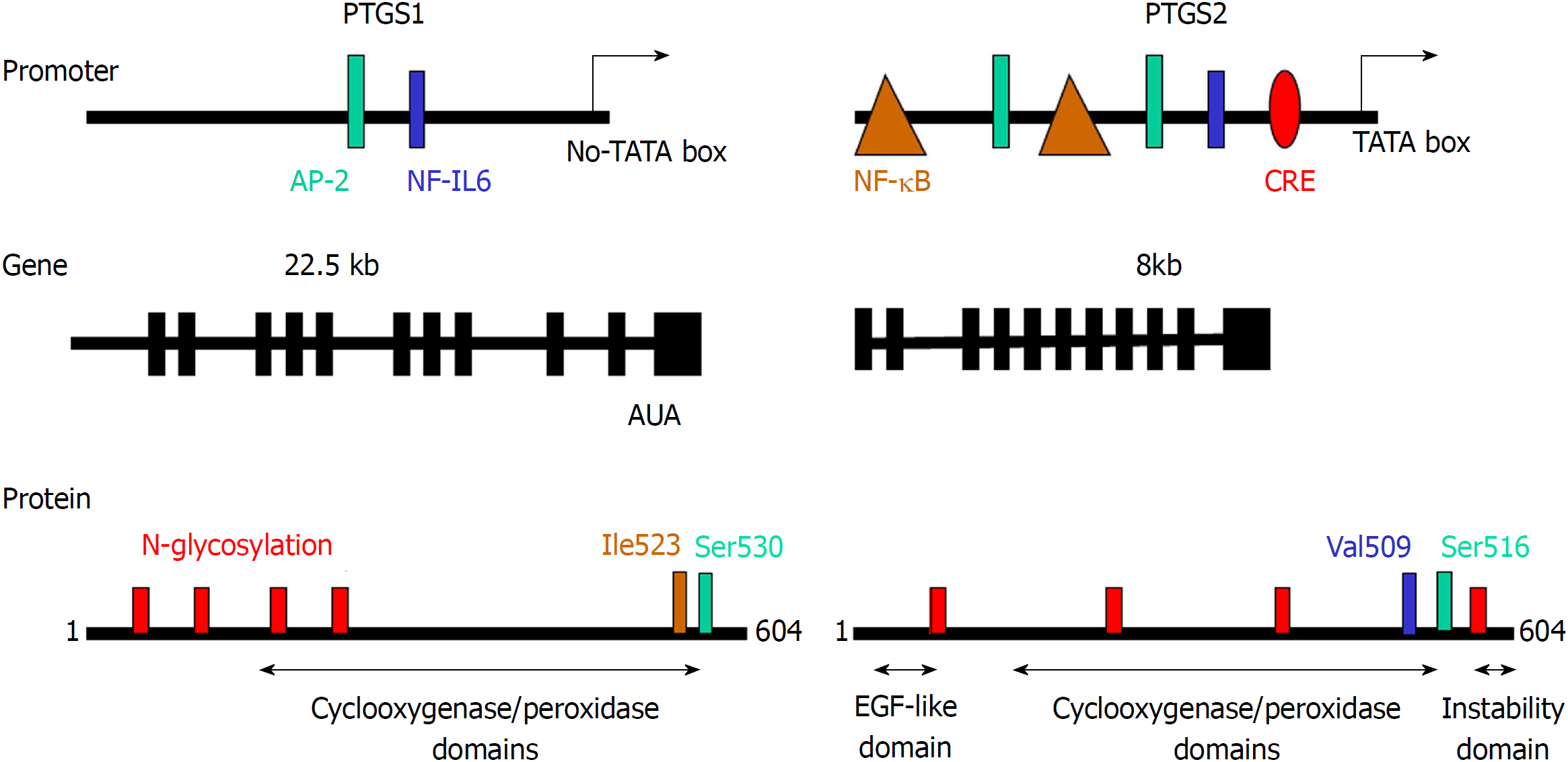
Figure 2 Comparison of the promoter region, gene structure and protein sequence of consensus prostaglandin-endoperoxide synthase 1 and 2.
The scheme shows the main transcription factors involved in the transcriptional control of prostaglandin-endoperoxide synthase (PTGS), the structure of the mRNA and the structural motifs present in the protein and relevant for the activity of the enzyme. The glycosylation of PTGS has effects on the activity and fate of the protein, protecting PTGS2 from proteasomal degradation. In addition to this, the electrophoretic mobility of the protein is altered by the glycosylation status of the protein. PTGS: Prostaglandin-endoperoxide synthase; AP-2: Activating protein 2; CRE: cAMP response element; NF-κB: Nuclear factor kappa B.
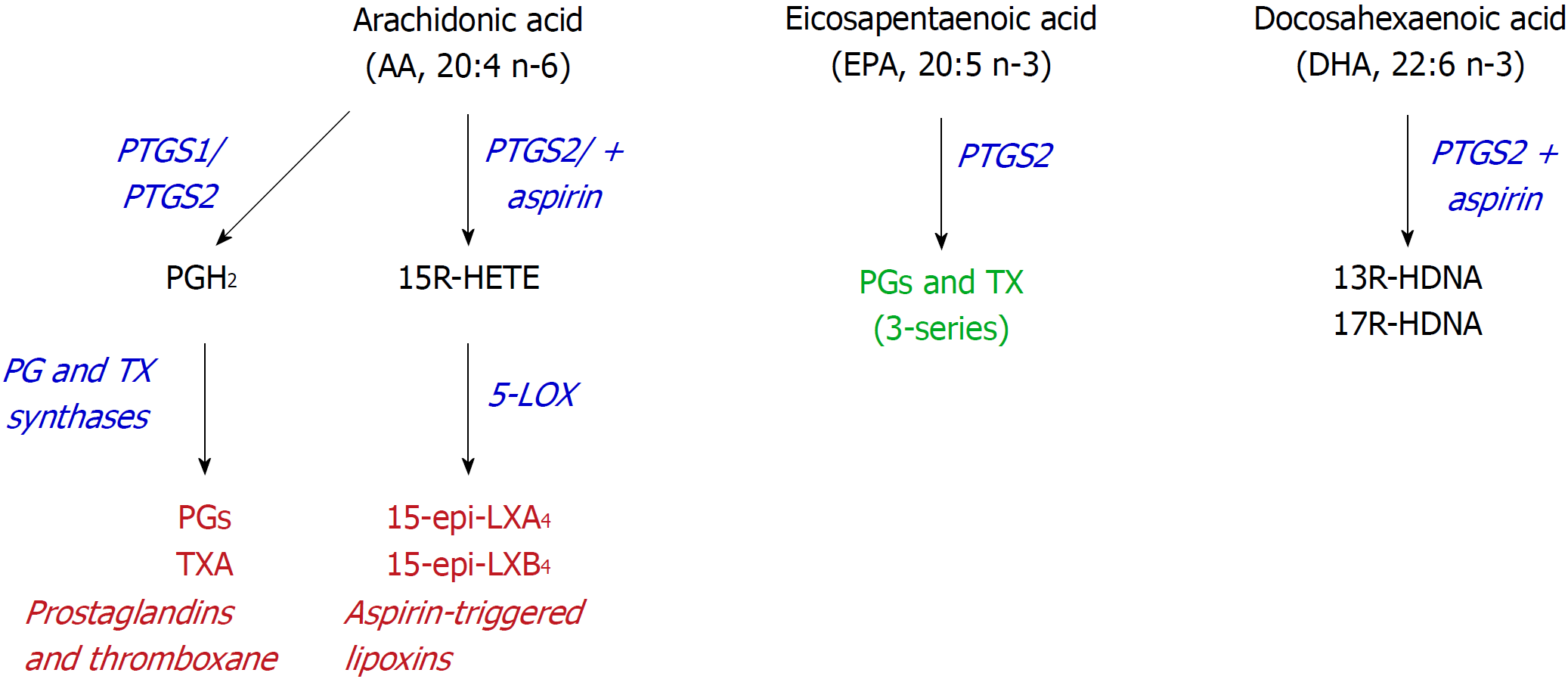
Figure 3 Alternative use of polyunsaturated fatty acids.
Aspirin may alter the activity of prostaglandin-endoperoxide synthase (PTGS) since it inhibits PTGS1, but retains the cyclooxygenase activity from PTGS2, leading to the synthesis of 15R-hydroxyeicosatetraenoic acid, from arachidonic acid, or 13- or 17R-hydroxy-docosahexaenoic acid, from docosahexaenoic acid. PTGS: Prostaglandin-endoperoxide synthase; HETE: Hydroxyeicosatetraenoic acid; HDHA: Hydroxy-docosahexaenoic acid.
PTGS2 EXPRESSION IN COLORECTAL CANCER TUMORIGENESIS
The role of PTGS2 expression in the initiation and progress of colorectal cancer (CRC) still remains a matter of debate. On one hand, epidemiological studies using broad (aspirin, indomethacin) or selective inhibitors of the PTGS isoforms at least suggest that under these conditions, prevention occurs in the development of CRC[1,16,31,34,35]. However, direct measurement of PGE2 levels in samples from adenomatous vs healthy tissue fails to show a clear cut-off supporting tumor growth and survival. In addition to this, the use of selective inhibitors of the EP receptors also contributes to the suggestion that autocrine signaling is perhaps critical in the commitment of the tumor cells to proliferate and invade the tissue via activation of mitogenic and metastatic pathways[6,27,31,34,36,37]. In addition to this, it is well known the capacity of PGE2 to favor angiogenesis of epithelial cells, contributing to the spreading and survival of the tumor. Moreover, due to the immunosuppressive activity of extracellular prostanoids, the anti-tumor role of the immune system is compromised, favoring the survival of the transformed cells in this microenvironment[3,33]. Not only the released products of PTGS2 have this capacity to alter cell fate, but at the intracellular level, prostaglandins itself or as result of oxidation due to increased oxidative stress may contribute to activate nuclear receptors, such as PPARs, that oppose to the pro-inflammatory defense mechanism favoring oncogenic progression[31,32]. Thus, the amount and fate of the products released by PTGS2 activity have different functions in the onset of CRC. Moreover, several authors have considered the possibility that, at least for PTGS1, it may exert “moonlighting” functions whose biological relevance remains to be established[38,39]. Additionally, the PTGS products can be modified by another series of enzymes, the 15PGDHs, which are transcriptionally regulated and determine the prostanoid levels coming from the PTGS activity, contributing in this way to the fine tuning of the activity of these lipid mediators and their involvement on the pathophysiology of CRC tumorigenesis[40,41]. Indeed, agreement exists in the opposite regulation of PTGS2 and 15PGDH in the sense that elevated PTGS2 levels repress 15PGDH expression and vice versa, elevated 15PGDH use to repress PTGS levels through different complementary mechanisms, involving destabilization of the PTGS2 mRNA via specific interaction with microRNAs[8,14,42]. Finally, the interplay between different contributors to CRC is moving towards a new integrative view that considers the immunological modulation due to several agents, including vitamin D, polyunsaturated fatty acids, diets with specific content in omega-3/-6, and pharmacological treatment with broad (aspirin) or selective PTGS2 inhibitors (coxibs). Together, these factors determine the immune modulation of what is defined as the immunomodulatory molecular pathological epidemiology (PME), as an integrated view of the environment-tumor-immune interactions, that may establish efficient protocols for immunoprevention and immunotherapy, leading to a better precision medicine[43,44].
PTGS2 is mainly regulated at the transcription level. The promoter region of PTGS2 contains several regulatory elements conferring response to transcription factors such as activating protein-2 (AP-2), nuclear factor kappa B (NF-κB), cAMP response element (CRE), E-box and Sp1, with various sites with impact on the promoter activity[35,45-47] (Figure 2). Despite this presence of canonical conserved transcription regulatory elements, preliminary data from our group show that the activity of the promoter has specific signatures when comparing its activity in rodent vs human cells, at least in the response to the engagement of NF-AT sites. Indeed, NF-AT inhibition with cyclosporine A or tacrolimus results in the repression of the pro-inflammatory transcriptional regulation in human cells, but not in the rodent counterparts, suggesting a specific fine-tuning of the promoter activity of this gene, at least in myeloid and hepatic cell lines (Figure 4). Moreover, PTGS2 expression is controlled also at post-transcriptional level; from a gene containing 10 exons and producing at least three products ranging from 4.6 to 2.8 kb, a regulatory site, positioned in the last exon that contains the 3’-UTR encoding sequences, is responsible for RNA instability[48]. As a result of this complex regulation, the levels of prostaglandins may vary significantly among several pathological situations, due to the availability of different substrates for the enzyme, the post-translational modifications occurring in a given tissue and/or the capacity to export and degrade the PTGS products (15PGDH; lipoxygenases, etc.). Indeed, it has been proposed a role for PGE2 in the CRC stem cell expansion and metastasis, at least in mice models of the disease[34]. Nevertheless, consensus exists regarding the fact that PTGS2 expression is associated with various pathophysiological events, ranging from inflammatory diseases to different cancers. The main problem encountered by researchers in the field is the frequent lack of correlation between mRNA and protein levels of PTGS2 and the corresponding biosynthesis of prostanoids, mainly PGE2. This is due to the exquisite control on the transcription of the gene; the existence of sequences that destabilize the mRNA and the posttranslational modifications that alter not only the catalytic activity of the enzyme, but also the stability of the protein, usually favoring proteasomal and non-proteasomal degradation[49-51]. Indeed, some suggestion of a ‘moonlighting’ effect for the protein has been reported for PTGS, an aspect of growing interest in the cancer field[38,39,52]. Finally, more than 40 clinical trials on the use of coxibs in CRC have been registered (ClinicalTrials.gov). Despite the controversy on the use of selective (coxibs) vs less specific PTGS2 inhibitors (NSAIDS) the main problems coming from these studies of chemoprevention are associated to the lack of specific biomarkers on the progress of the disease[53] and the existence of side effects that, at the end, represent a serious bias in the establishment of the critical parameters associated to the onset of the pathology[16,34,54].
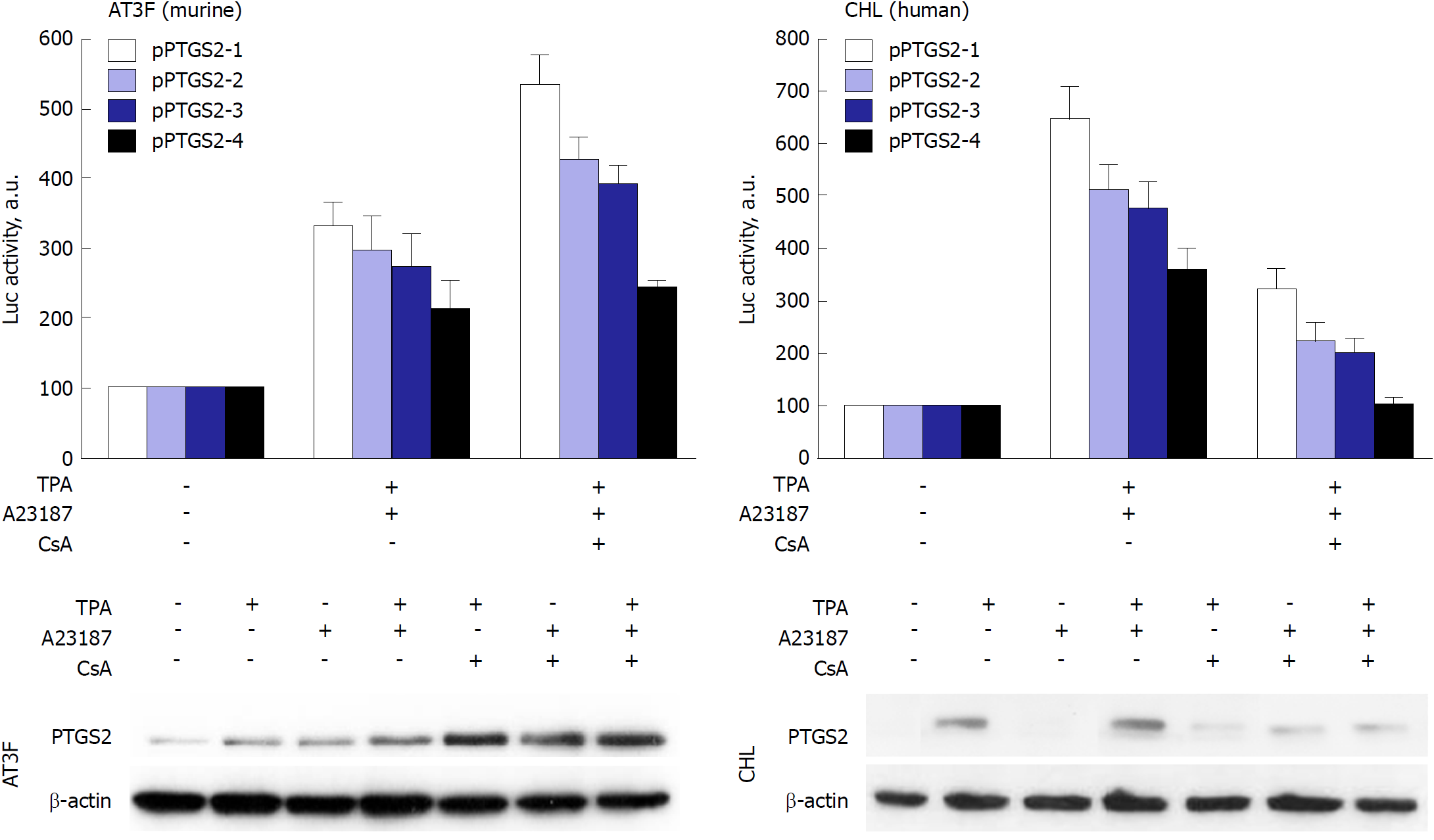
Figure 4 Prostaglandin-endoperoxide synthase 2 exhibits species-specific transcriptional control.
Despite the broad conservation of the transcription factor motifs in the prostaglandin-endoperoxide synthase 2 promoter, the activity of the promoter in response to the NF-AT inhibitor cyclosporine A (CsA) is repressed in human hepatic cell lines, but enhanced in murine hepatic counterparts. To confirm this effect, murine AT3F hepatic cells and human CHL hepatic cells were transfected with different constructions of the promoter linked to a luciferase reporter gene (see[46] for details). The protein levels and the luciferase activity were determined at 24h after treatment with the indicated stimuli: tetradecanoylphorbol acetate 100 nmol/L, Ca2+-ionophore A23187 1 μmol/L; CsA 100 nmol/L. TPA: Tetradecanoylphorbol acetate; CsA: Cyclosporine A; PTGS: Prostaglandin-endoperoxide synthase.
PATHOPHYSIOLOGICAL RELEVANCE OF THE POST-TRANSLATIONAL REGULATION OF PTGS2
Apart from the classic pharmacological acetylation by aspirin on Ser530 in PTGS1 and Ser516 in PTGS2[55], that prevents the full activity of the enzyme [PTGS2 retains the ability to generate 15R-hydroxyeicosatetraenoic acid (HETE) from arachidonic acid], other physiological post-translational modifications have been described for the PTGS isoenzymes. These modifications have an impact on the catalytic activity, on the subcellular localization, and on the targeting of the protein for degradation via proteasomal or non-proteasomal (endoplasmic reticulum-dependent pathways) pathways[51]. Overall, these modifications are in the basis of most of the pathophysiological responses associated to the different conditions in which PTGS2 is expressed.
Since the catalytic activity of PTGS requires a functional heme group in the protein and this can be modified by different oxidants, such as nitric oxide and other free radicals interacting with the prosthetic group, the enzymatic activity can be altered in this way[56]. This is especially relevant in situations in which the high throughput nitric oxide synthase (NOS-2) is expressed and a high synthesis of NO occurs; however, this cannot exclude other NOS isoenzymes that, although releasing lesser NO, because of the proximity to PTGS may selectively affect its activity, although discrepancies in the literature exist describing either inhibition or activation by NO. In fact, a complex crosstalk between the NOS and the PTGS systems has been reported in spheroid cultures of CRC: NO inhibits PTGS activity through different pathways, including S-nitrosylation by peroxynitrite[57].
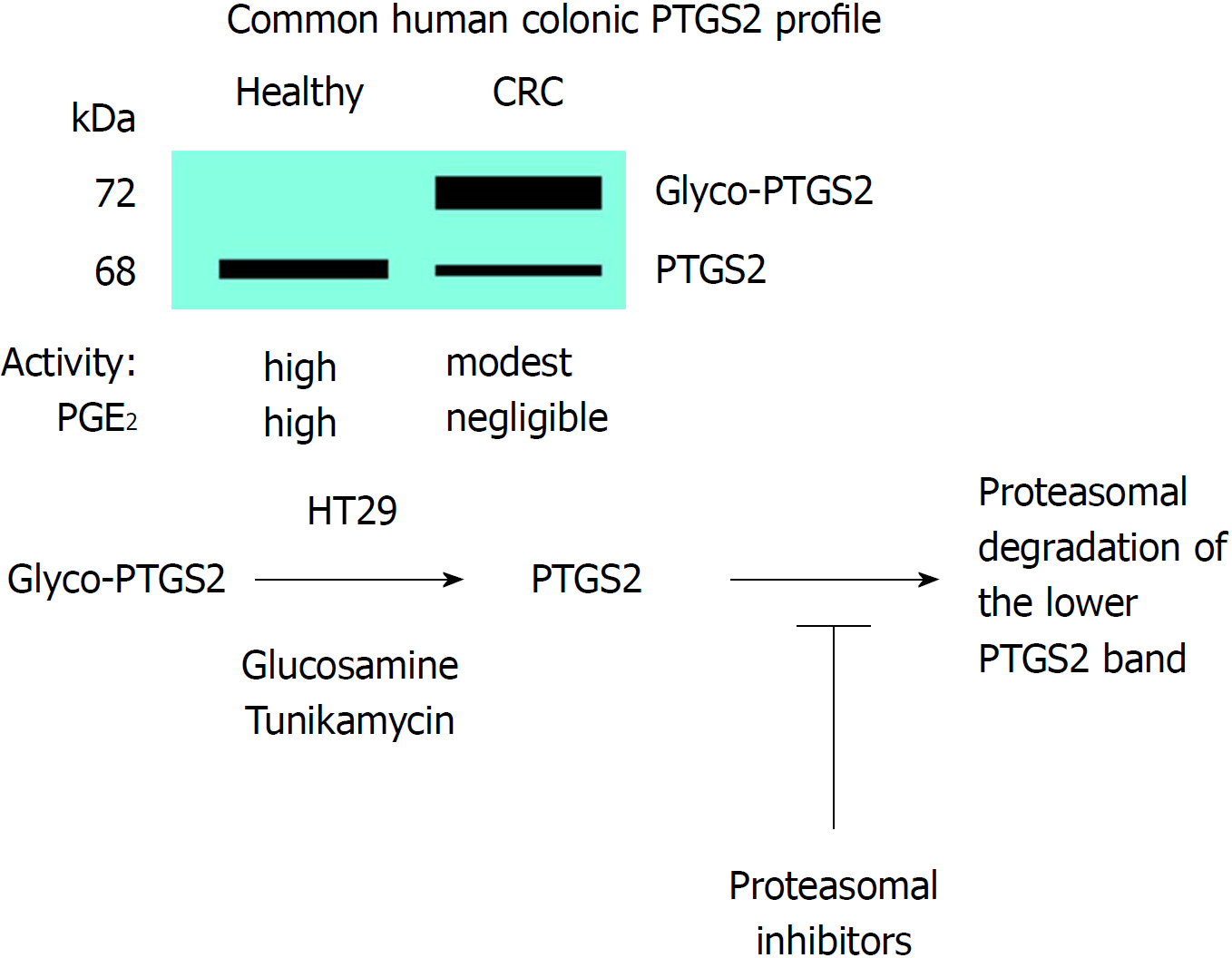
Western blot analysis of PTGS2 in samples of CRC patients and in tumor colonic cell lines have evidenced the presence of several immunodetected bands corresponding to PTGS2. The change in electrophoretic mobility of the proteins is due to the presence of glycosylated motifs in the protein as confirmed by biochemical and proteomic studies. Indeed, proteomic analysis of PTGS2 characterized the main glycosylated aminoacidic residues[58]. Both N-and O-linked glycosylation in asparagine and serine/threonine are possible. This glycosylated form is prevalent in CRC samples and in tumor colonic cell lines, but is usually absent in samples of healthy colonic tissue when PTGS2 is expressed[21,59-61]. Indeed, this glycosylated PTGS2 is more resistant to proteolytic degradation than the non-glycosylated counterpart; however, we have been unable to identify the specific glycosylation(s) of this PTGS2 by using proteomic approaches. In fact, this is surprising since previous work identified the N-glycosylation at Asn580/594 as a condition favoring its proteolysis through the ER-dependent pathway[51]. However, the absence of glycosyl marks in the protein, after treatment with glucosamine that reverts the glycosylated band to the non-glycosylated form, targets the protein for a rapid degradation, as reflected by the absence of changes in the mRNA levels, but inducing a significant decrease in the protein levels of HT29 cells treated with glucosamine[61]. Moreover, inhibitors of the proteasome prevent the degradation of PTGS2 in colonic cell lines treated with glucosamine. In addition to this, the catalytic activity of glycosylated PTGS2 is lesser than the corresponding to the non-glycosylated form[59,61]. Finally, additional work is required to address the effect of PTGS2 glycosylation on the response to the classic inhibitors of the enzyme, a condition of pharmacologic and therapeutic interest in the regulation of the catalytic activity of the enzyme[62]. Figure 5 summarizes the effect of glycosylation on PTGS2 activity and fate.
Figure 5 Glycosylation of prostaglandin-endoperoxide synthase 2 in colorectal cancer.
The main form in colorectal cancer tissues is the glycosylated 72 kDa protein. This glycosylated prostaglandin-endoperoxide synthase 2 (PTGS2) is also expressed in several tumor colonic cell lines and can be deglycosylated after incubation of the cells with glucosamine or tunikamycin. After this treatment, PTGS2 is rapidly degraded via proteasomal activity. CRC: Colorectal cancer; PTGS: Prostaglandin-endoperoxide synthase.
Another issue no completely resolved in the post-translational modifications of PTGS2 is the phosphorylation in tyrosine and serine/threonine residues. In fact, PTGS2 contains consensus motifs for the phosphorylation by protein tyrosine kinases, such as FYN. However, direct proofs for the occurring of such phosphorylations have failed to provide sufficient evidence. The same happens for the PKC phosphorylation motifs present in the protein. However, it appears that specific tyrosine phosphorylation is required for the functional glycosylation of PTGS2, suggesting the convergence of different pathways in the final post-translational modifications of the enzyme, with relevance not only for the enzymatic activity but also for the targeting and degradation[9].
CONCLUSION
Understanding the pathophysiological role of the post-translational modifications of PTGS2 remains a subject of research in the area of oncology. Assessment of the role of prostanoids in CRC initiation and progression may contribute to a better management of the patients and in the proposal of therapeutic interventions intended to regulate colonic PTGS2 activity. Finally, the possibility exists to use PTGS2 post-translational modifications in biopsies as an additional predictive biomarker in CRC evaluation, and a better integration in the immunomodulatory-molecular pathological epidemiology[43].
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): A
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chan AT S- Editor: Ma RY L- Editor: A E- Editor: Yin SY